Software
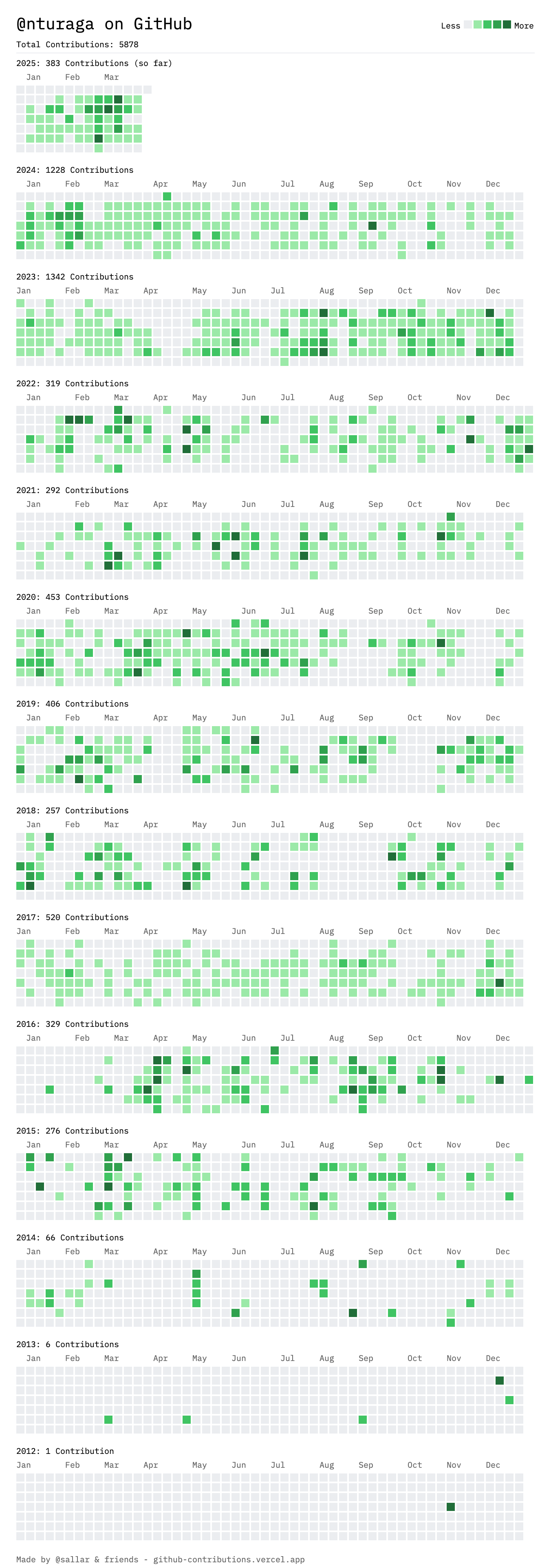
Overall Github contribution up to March 2025
Open Source Software in R
Some of the R packages i’ve written are listed below:
-
BiocKubeInstall: Bioconductor package used to create binaries for docker images produced by Bioconductor. The package installation and binary creation is parallelized on a Kubernetes cluster launched (at the moment) using Google Kubernetes Engine(GKE) and Azure (AKS).
-
AnVIL: The AnVIL R-package provides end-user and developer functionality for AnVIL cloud computing resource.
-
BiocParallel: The BiocParallel package provides modified versions and novel implementation of functions for parallel evaluation, tailored to use with Bioconductor objects. I implemented the BatchtoolsParam interface, which allows distributed computation on different job scheduling systems on HPC.
-
BiocDockerManager: The BiocDockerManager package allows the installation and management of docker images provided by the Bioconductor project. A convenient package to install images, update images, and find which Bioconductor based docker images are available.
-
AWSParallel: The AWSParallel package provides facilities to perform parallel evaluation with AWS infrastructure. It connects with BiocParallel and works with Bioconductor objects. The package has a comprehensive vignette which shows how to execute batch jobs.
This package is not in Bioconductor but remains available on Github because it uses StarCluster, which is outdated for AWS cluster management.
Open Source Docker Images
I create, design, and maintain docker images which have an interactive environment as a front end (Jupyter or RStudio). This is part of my work in scalable cloud computing.
Some of the images I produce are listed here. These are production level docker images that serve multiple purposes, from usage on local machines, cloud services, and HPC systems. I’ve removed legacy images that I don’t maintain anymore. Please note that this is not an exhaustive list.
Bioconductor Images
-
bioconductor/bioconductor_docker
This image can install almost all the packages in Bioconductor (>99.8%). It’s one of the most widely used images for Bioconductor. It operates on an RStudio front end. The image has multiple versions, one for each release and the latest devel image, which is updated weekly through a custom Github action.
-
bioconductor/bioconductor_full (deprecated 2019)
This image was deprecated because the inheritance chain of docker images became very cumbersome to maintain. The deprecation was in favor of
bioconductor/bioconductor_docker.NOTE: Some of the other legacy images that I maintained are listed on here. These are now deprecated.
AnVIL images
The AnVIL project allows large scale genomic data analysis on the Google Cloud. The following images are custom built for the AnVIL project and are on the Terra application. The images here have customization to access workspaces and files from AnVIL. These images are on the Google container registry.
-
anvil-docker/anvil-rstudio-base
Launch RStudio Community edition as an interactive environment on the AnVIL platform using this image.
-
anvil-docker/anvil-rstudio-bioconductor
Launch RStudio Community edition as an interactive environment with pre-installed Bioconductor packages on the AnVIL platform using this image.
-
terra-docker/terra-jupyter-r
Launch a Jupyter notebook as an interactive environment with an R kernel the AnVIL platform using this image.
-
terra-docker/terra-jupyter-bioconductor
Launch a Jupyter notebook as an interactive environment with an R kernel and pre-installed Bioconductor packages on the AnVIL platform using this image.
-
terra-docker/rstudio-pro
Launch an RStudio pro server as an interactive environment on the AnVIL platform using this image.
Open Source Software in Python
Python is my favorite language along with R. I implement many infrastructure related scripts in Python, which unfortunately do not make “package” format.
-
Git resources in Python: The
bioc_git_transitionpackage was initially used to transition all of Bioconductor from SVN to Git. The package has a considerable number ofgitsubprocess commands, which had to be run during the time of transition in 2017 but stopped after. -
Bioconductor git hooks: The git hooks that Bioconductor’s git server uses on each package are in this git repository.
-
galaxyproject/planemo: Command-line utilities to assist in developing Galaxy tools. I specifically contributed to planemo by implementing features to add Bioconductor packages as Galaxy tools.
-
bioaRchive: bioaRchive stores versions of Bioconductor packages to promote interoperability between Galaxy and Bioconductor. The main goal is to allow users access to all the versions of current BioConductor packages.